Recent advancements in understanding protein-protein interactions within multicellular organisms have been propelled by a groundbreaking approach developed by a collaborative research team at The University of Hong Kong (HKU). Led by Professor Xiang David Li from the Department of Chemistry and Professor Chaogu Zheng from the School of Biological Sciences, along with Dr. Xiucong Bao from the School of Biomedical Sciences, the team introduced the Methionine Analog-based Cell-Specific Proteomics and Interactomics (MACSPI) method. This innovative technique enables the labeling of proteins in specific cells and the capture of their interactions, providing valuable insights into cellular functions within living organisms.
The complexity of multicellular organisms, such as animals and plants, stems from the diverse functions of their complex cells. These cells rely on intricate protein interactions to execute specific tasks and form complex molecular machinery. However, conventional studies on protein-protein interactions often lack cellular context as they are typically conducted in vitro or using isolated cells. Consequently, methods to investigate these interactions in a tissue-specific manner have been largely absent.
To address this technological gap, the HKU research team devised a chemical biology approach utilizing a bifunctional amino acid probe to label proteins from specific cells. This probe facilitates the isolation of labeled proteins and captures protein-protein interactions through photo-crosslinking. By employing this method, the team identified numerous tissue-specific proteins and their interactions, enhancing our understanding of cellular mechanisms within living organisms and providing insights into various biological phenomena, including organ development and disease pathogenesis. The research findings were recently published in the esteemed multidisciplinary journal Proceedings of the National Academy of Sciences (PNAS).
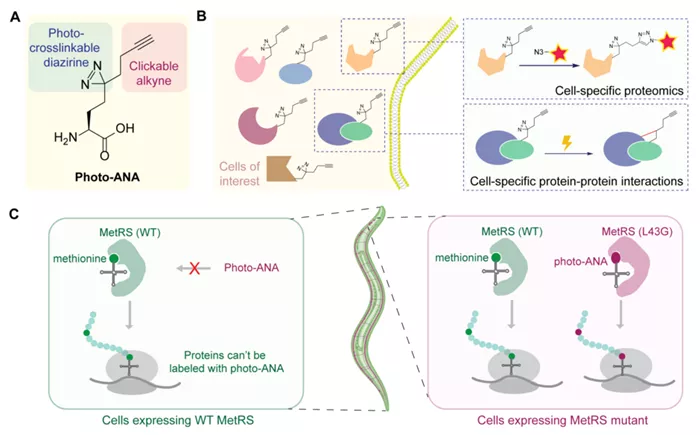
The innovative design of MACSPI centers around the development of an unnatural amino acid, termed photo-ANA, structurally resembling methionine but featuring additional components. These components include an alkyne group for chemical handling of labeled proteins and a diazirine group for light-induced creation of stable covalent linkages between labeled proteins and their interacting molecules. Furthermore, the team engineered an enzyme, MetRS, capable of recognizing and incorporating the unnatural amino acid into proteins during their synthesis. By controlling the expression of this engineered enzyme in specific tissues, only proteins from the tissue of interest are labeled by the chemical probe. Additionally, light-induced crosslinking enables the capture and isolation of protein complexes from specific tissues.
In a proof-of-concept demonstration, the researchers applied the MACSPI method to profile proteins from muscle cells and neurons in the model organism Caenorhabditis elegans, uncovering numerous novel tissue-specific proteins. Moreover, the method proved effective in capturing tissue-specific protein-protein interactions, as evidenced by the identification of tissue-specific interactors of a ubiquitously expressed protein, HSP90, a molecular chaperone. The study revealed that HSP90 binds to distinct sets of proteins, regulating different biological processes in muscles and neurons.
Professor Xiang David Li emphasized the significance of innovative chemical labeling methods in addressing complex biological inquiries. Professor Chaogu Zheng highlighted the potential of MACSPI in unraveling the molecular mechanisms underlying pathological processes, such as neurodegeneration in Parkinson’s disease models.
The research team envisions that the MACSPI method will find widespread applications in profiling proteomes and interactomes with spatial and temporal specificity across diverse multicellular organisms, facilitating a wide array of biological and biomedical research endeavors.
The study represents a collaborative effort among Professor Xiang David Li’s team at the Department of Chemistry, Professor Chaogu Zheng’s team at the School of Biological Sciences, and Dr. Xiucong Bao’s team at the School of Biomedical Sciences at HKU. Dr. Siyue Huang from Professor Li’s team and Ms. Qiao Ran from Professor Zheng’s team serve as co-first authors of the study. Initial support for the research was provided by seed funds from the Strategic Interdisciplinary Research Scheme at HKU, followed by grants from various funding agencies including the Hong Kong Research Grants Council Collaborative Research Fund, the Areas of Excellence Scheme, the General Research Fund, Food and Health Bureau, and the National Natural Science Foundation of China.
The research paper titled “MACSPI Enables Tissue-Selective Proteomic and Interactomic Analysis in Multicellular Organisms” was published in the Proceedings of the National Academy of Sciences (PNAS) on May 13, 2024.